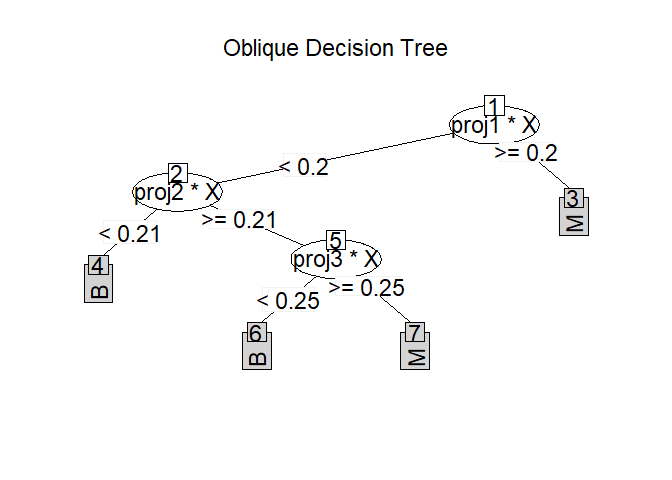
Classification and regression using an oblique decision tree (ODT) in which each node is split by a linear combination of predictors. Different methods are provided for selecting the linear combinations, while the splitting values are chosen by one of three criteria.
Usage
ODT(X, ...)
# S3 method for class 'formula'
ODT(
formula,
data = NULL,
Xsplit = NULL,
split = "auto",
lambda = "log",
NodeRotateFun = "RotMatPPO",
FunDir = getwd(),
paramList = NULL,
glmnetParList = NULL,
MaxDepth = Inf,
numNode = Inf,
MinLeaf = 10,
Levels = NULL,
subset = NULL,
weights = NULL,
na.action = na.fail,
catLabel = NULL,
Xcat = 0,
Xscale = "Min-max",
TreeRandRotate = FALSE,
...
)
# Default S3 method
ODT(
X,
y,
Xsplit = NULL,
split = "auto",
lambda = "log",
NodeRotateFun = "RotMatPPO",
FunDir = getwd(),
paramList = NULL,
glmnetParList = NULL,
MaxDepth = Inf,
numNode = Inf,
MinLeaf = 10,
Levels = NULL,
subset = NULL,
weights = NULL,
na.action = na.fail,
catLabel = NULL,
Xcat = 0,
Xscale = "Min-max",
TreeRandRotate = FALSE,
...
)Arguments
- X
An n by d numeric matrix (preferable) or data frame.
- ...
Optional parameters to be passed to the low level function.
- formula
Object of class
formulawith a response describing the model to fit. If this is a data frame, it is taken as the model frame. (seemodel.frame)- data
Training data of class
data.framecontaining variables named in the formula. Ifdatais missing it is obtained from the current environment byformula.- Xsplit
Splitting variables used to construct linear model trees. The default value is NULL and is only valid when split="linear".
- split
The criterion used for splitting the nodes. "entropy": information gain and "gini": gini impurity index for classification; "mse": mean square error for regression; "linear": mean square error for linear model. 'auto' (default): If the response in
dataoryis a factor, "gini" is used, otherwise "mse" is assumed.- lambda
The argument of
splitis used to determine the penalty level of the partition criterion. Three options are provided including,lambda=0: no penalty;lambda=2: AIC penalty;lambda='log'(Default): BIC penalty. In Addition, lambda can be any value from 0 to n (training set size).- NodeRotateFun
Name of the function of class
characterthat implements a linear combination of predictors in the split node. including"RotMatPPO": projection pursuit optimization model (
PPO), seeRotMatPPO(default, model="PPR")."RotMatRF": single feature similar to CART, see
RotMatRF."RotMatRand": random rotation, see
RotMatRand."RotMatMake": users can define this function, for details see
RotMatMake.
- FunDir
The path to the
functionof the user-definedNodeRotateFun(default current working directory).- paramList
List of parameters used by the functions
NodeRotateFun. If left unchanged, default values will be used, for details seedefaults.- glmnetParList
List of parameters used by the functions
glmnetandcv.glmnetin packageglmnet.glmnetParList=list(lambda = 0)is Ordinary Least Squares (OLS) regression,glmnetParList=list(family = "gaussian")(default) is regression model andglmnetParList=list(family = "binomial" or "multinomial")is classification model. If left unchanged, default values will be used, for details seeglmnetandcv.glmnet.- MaxDepth
The maximum depth of the tree (default
Inf).- numNode
Number of nodes that can be used by the tree (default
Inf).- MinLeaf
Minimal node size (Default 10).
- Levels
The category label of the response variable when
splitis not equal to 'mse'.- subset
An index vector indicating which rows should be used. (NOTE: If given, this argument must be named.)
- weights
Vector of non-negative observational weights; fractional weights are allowed (default NULL).
- na.action
A function to specify the action to be taken if NAs are found. (NOTE: If given, this argument must be named.)
- catLabel
A category labels of class
listin predictors. (default NULL, for details see Examples)- Xcat
A class
vectoris used to indicate which predictor is the categorical variable. The default Xcat=0 means that no special treatment is given to category variables. When Xcat=NULL, the predictor x that satisfies the condition "(length(table(x))<10) & (length(x)>20)" is judged to be a category variable.- Xscale
Predictor standardization methods. " Min-max" (default), "Quantile", "No" denote Min-max transformation, Quantile transformation and No transformation respectively.
- TreeRandRotate
If or not to randomly rotate the training data before building the tree (default FALSE, see
RandRot).- y
A response vector of length n.
Value
An object of class ODT containing a list of components::
call: The original call to ODT.terms: An object of classc("terms", "formula")(seeterms.object) summarizing the formula. Used by various methods, but typically not of direct relevance to users.split,LevelsandNodeRotateFunare important parameters for building the tree.predicted: the predicted values of the training data.projections: Projection direction for each split node.paramList: Parameters in a named list to be used byNodeRotateFun.data: The list of data related parameters used to build the tree.tree: The list of tree related parameters used to build the tree.structure: A set of tree structure data records.nodeRotaMat: Record the split variables (first column), split node serial number (second column) and rotation direction (third column) for each node. (The first column and the third column are 0 means leaf nodes)nodeNumLabel: Record each leaf node's category for classification or predicted value for regression (second column is data size). (Each column is 0 means it is not a leaf node)nodeCutValue: Record the split point of each node. (0 means leaf nodes)nodeCutIndex: Record the index values of the partitioning variables selected based on the partition criterionsplit.childNode: Record the number of child nodes after each splitting.nodeDepth: Record the depth of the tree where each node is located.nodeIndex: Record the indices of the data used in each node.glmnetFit: Record the model fitted by functionglmnetused in each node.
References
Zhan, H., Liu, Y., & Xia, Y. (2022). Consistency of The Oblique Decision Tree and Its Random Forest. arXiv preprint arXiv:2211.12653.
Examples
# Classification with Oblique Decision Tree.
data(seeds)
set.seed(221212)
train <- sample(1:209, 100)
train_data <- data.frame(seeds[train, ])
test_data <- data.frame(seeds[-train, ])
tree <- ODT(varieties_of_wheat ~ ., train_data, split = "entropy")
pred <- predict(tree, test_data[, -8])
# classification error
(mean(pred != test_data[, 8]))
#> [1] 0.09174312
# Regression with Oblique Decision Tree.
data(body_fat)
set.seed(221212)
train <- sample(1:252, 100)
train_data <- data.frame(body_fat[train, ])
test_data <- data.frame(body_fat[-train, ])
tree <- ODT(Density ~ ., train_data,
split = "mse",
NodeRotateFun = "RotMatPPO", paramList = list(model = "Log", dimProj = "Rand")
)
pred <- predict(tree, test_data[, -1])
# estimation error
mean((pred - test_data[, 1])^2)
#> [1] 0.0005496953
# Use "Z" as the splitting variable to build a linear model tree for "X" and "y".
set.seed(10)
cutpoint <- 50
X <- matrix(rnorm(100 * 10), 100, 10)
age <- sample(seq(20, 80), 100, replace = TRUE)
height <- sample(seq(50, 200), 100, replace = TRUE)
weight <- sample(seq(5, 150), 100, replace = TRUE)
Z <- cbind(age = age, height = height, weight = weight)
mu <- rep(0, 100)
mu[age <= cutpoint] <- X[age <= cutpoint, 1] + X[age <= cutpoint, 2]
mu[age > cutpoint] <- X[age > cutpoint, 1] + X[age > cutpoint, 3]
y <- mu + rnorm(100)
# Regression model tree
my.tree <- ODT(
X = X, y = y, Xsplit = Z, split = "linear", lambda = 0,
NodeRotateFun = "RotMatRF",
glmnetParList = list(lambda = 0, family = "gaussian")
)
pred <- predict(my.tree, X, Xsplit = Z)
# fitting error
mean((pred - y)^2)
#> [1] 0.9035932
mean((my.tree$predicted - y)^2)
#> [1] 0.9035932
# Classification model tree
y1 <- (y > 0) * 1
my.tree <- ODT(
X = X, y = y1, Xsplit = Z, split = "linear", lambda = 0,
NodeRotateFun = "RotMatRF", MinLeaf = 10, MaxDepth = 5,
glmnetParList = list(family = "binomial")
)
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
#> Warning: one multinomial or binomial class has fewer than 8 observations; dangerous ground
(class <- predict(my.tree, X, Xsplit = Z, type = "pred"))
#> Warning: number of rows of result is not a multiple of vector length (arg 1)
#> [1] "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0"
#> [19] "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0"
#> [37] "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0"
#> [55] "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0"
#> [73] "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0" "0"
#> [91] "0" "0" "0" "0" "0" "0" "0" "0" "0" "0"
(prob <- predict(my.tree, X, Xsplit = Z, type = "prob"))
#> Warning: number of rows of result is not a multiple of vector length (arg 1)
#> 0 1
#> [1,] 1 0
#> [2,] 1 0
#> [3,] 1 0
#> [4,] 1 0
#> [5,] 1 0
#> [6,] 1 0
#> [7,] 1 0
#> [8,] 1 0
#> [9,] 1 0
#> [10,] 1 0
#> [11,] 1 0
#> [12,] 1 0
#> [13,] 1 0
#> [14,] 1 0
#> [15,] 1 0
#> [16,] 1 0
#> [17,] 1 0
#> [18,] 1 0
#> [19,] 1 0
#> [20,] 1 0
#> [21,] 1 0
#> [22,] 1 0
#> [23,] 1 0
#> [24,] 1 0
#> [25,] 1 0
#> [26,] 1 0
#> [27,] 1 0
#> [28,] 1 0
#> [29,] 1 0
#> [30,] 1 0
#> [31,] 1 0
#> [32,] 1 0
#> [33,] 1 0
#> [34,] 1 0
#> [35,] 1 0
#> [36,] 1 0
#> [37,] 1 0
#> [38,] 1 0
#> [39,] 1 0
#> [40,] 1 0
#> [41,] 1 0
#> [42,] 1 0
#> [43,] 1 0
#> [44,] 1 0
#> [45,] 1 0
#> [46,] 1 0
#> [47,] 1 0
#> [48,] 1 0
#> [49,] 1 0
#> [50,] 1 0
#> [51,] 1 0
#> [52,] 1 0
#> [53,] 1 0
#> [54,] 1 0
#> [55,] 1 0
#> [56,] 1 0
#> [57,] 1 0
#> [58,] 1 0
#> [59,] 1 0
#> [60,] 1 0
#> [61,] 1 0
#> [62,] 1 0
#> [63,] 1 0
#> [64,] 1 0
#> [65,] 1 0
#> [66,] 1 0
#> [67,] 1 0
#> [68,] 1 0
#> [69,] 1 0
#> [70,] 1 0
#> [71,] 1 0
#> [72,] 1 0
#> [73,] 1 0
#> [74,] 1 0
#> [75,] 1 0
#> [76,] 1 0
#> [77,] 1 0
#> [78,] 1 0
#> [79,] 1 0
#> [80,] 1 0
#> [81,] 1 0
#> [82,] 1 0
#> [83,] 1 0
#> [84,] 1 0
#> [85,] 1 0
#> [86,] 1 0
#> [87,] 1 0
#> [88,] 1 0
#> [89,] 1 0
#> [90,] 1 0
#> [91,] 1 0
#> [92,] 1 0
#> [93,] 1 0
#> [94,] 1 0
#> [95,] 1 0
#> [96,] 1 0
#> [97,] 1 0
#> [98,] 1 0
#> [99,] 1 0
#> [100,] 1 0
# Projection analysis of the oblique decision tree.
data(iris)
tree <- ODT(Species ~ .,
data = iris, split = "gini",
paramList = list(model = "PPR", numProj = 1)
)
print(round(tree[["projections"]], 3))
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> proj1 -0.167 -0.215 0.763 0.587
#> proj2 -0.087 -0.223 0.676 0.697
### Train ODT on one-of-K encoded categorical data ###
# Note that the category variable must be placed at the beginning of the predictor X
# as in the following example.
set.seed(22)
Xcol1 <- sample(c("A", "B", "C"), 100, replace = TRUE)
Xcol2 <- sample(c("1", "2", "3", "4", "5"), 100, replace = TRUE)
Xcon <- matrix(rnorm(100 * 3), 100, 3)
X <- data.frame(Xcol1, Xcol2, Xcon)
Xcat <- c(1, 2)
catLabel <- NULL
y <- as.factor(sample(c(0, 1), 100, replace = TRUE))
tree <- ODT(X, y, split = "entropy", Xcat = NULL)
#> Warning: The categorical variable 1, 2 has been transformed into an one-of-K encode variables!
head(X)
#> Xcol1 Xcol2 X1 X2 X3
#> 1 B 5 -0.04178453 2.3962339 -0.01443979
#> 2 A 4 -1.66084623 -0.4397486 0.57251733
#> 3 B 2 -0.57973333 -0.2878683 1.24475578
#> 4 B 1 -0.82075051 1.3702900 0.01716528
#> 5 C 5 -0.76337897 -0.9620213 0.25846351
#> 6 A 5 -0.37720294 -0.1853976 1.04872159
#> Xcol1 Xcol2 X1 X2 X3
#> 1 B 5 -0.04178453 2.3962339 -0.01443979
#> 2 A 4 -1.66084623 -0.4397486 0.57251733
#> 3 B 2 -0.57973333 -0.2878683 1.24475578
#> 4 B 1 -0.82075051 1.3702900 0.01716528
#> 5 C 5 -0.76337897 -0.9620213 0.25846351
#> 6 A 5 -0.37720294 -0.1853976 1.04872159
# one-of-K encode each categorical feature and store in X1
numCat <- apply(X[, Xcat, drop = FALSE], 2, function(x) length(unique(x)))
# initialize training data matrix X
X1 <- matrix(0, nrow = nrow(X), ncol = sum(numCat))
catLabel <- vector("list", length(Xcat))
names(catLabel) <- colnames(X)[Xcat]
col.idx <- 0L
# convert categorical feature to K dummy variables
for (j in seq_along(Xcat)) {
catMap <- (col.idx + 1):(col.idx + numCat[j])
catLabel[[j]] <- levels(as.factor(X[, Xcat[j]]))
X1[, catMap] <- (matrix(X[, Xcat[j]], nrow(X), numCat[j]) ==
matrix(catLabel[[j]], nrow(X), numCat[j], byrow = TRUE)) + 0
col.idx <- col.idx + numCat[j]
}
X <- cbind(X1, X[, -Xcat])
colnames(X) <- c(paste(rep(seq_along(numCat), numCat), unlist(catLabel),
sep = "."
), "X1", "X2", "X3")
# Print the result after processing of category variables.
head(X)
#> 1.A 1.B 1.C 2.1 2.2 2.3 2.4 2.5 X1 X2 X3
#> 1 0 1 0 0 0 0 0 1 -0.04178453 2.3962339 -0.01443979
#> 2 1 0 0 0 0 0 1 0 -1.66084623 -0.4397486 0.57251733
#> 3 0 1 0 0 1 0 0 0 -0.57973333 -0.2878683 1.24475578
#> 4 0 1 0 1 0 0 0 0 -0.82075051 1.3702900 0.01716528
#> 5 0 0 1 0 0 0 0 1 -0.76337897 -0.9620213 0.25846351
#> 6 1 0 0 0 0 0 0 1 -0.37720294 -0.1853976 1.04872159
#> 1.A 1.B 1.C 2.1 2.2 2.3 2.4 2.5 X1 X2 X3
#> 1 0 1 0 0 0 0 0 1 -0.04178453 2.3962339 -0.01443979
#> 2 1 0 0 0 0 0 1 0 -1.66084623 -0.4397486 0.57251733
#> 3 0 1 0 0 1 0 0 0 -0.57973333 -0.2878683 1.24475578
#> 4 0 1 0 1 0 0 0 0 -0.82075051 1.3702900 0.01716528
#> 5 0 0 1 0 0 0 0 1 -0.76337897 -0.9620213 0.25846351
#> 6 1 0 0 0 0 0 0 1 -0.37720294 -0.1853976 1.04872159
catLabel
#> $Xcol1
#> [1] "A" "B" "C"
#>
#> $Xcol2
#> [1] "1" "2" "3" "4" "5"
#>
#> $Xcol1
#> [1] "A" "B" "C"
#>
#> $Xcol2
#> [1] "1" "2" "3" "4" "5"
tree <- ODT(X, y, split = "gini", Xcat = c(1, 2), catLabel = catLabel, NodeRotateFun = "RotMatRF")