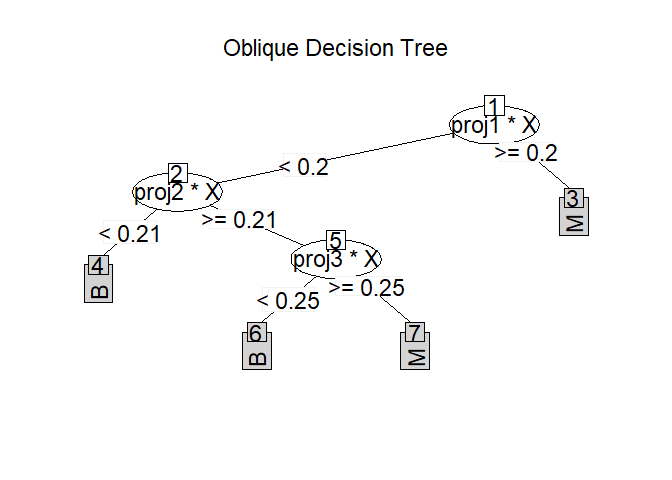
Print the oblique decision tree structure.
Usage
# S3 method for class 'ODT'
print(x, projection = FALSE, cutvalue = FALSE, verbose = TRUE, ...)Arguments
- x
An object of class
ODT.- projection
Print projection coefficients in each node if TRUE.
- cutvalue
Print cutoff values in each node if TRUE.
- verbose
Print if TRUE, no output if FALSE.
- ...
Arguments to be passed to methods.
References
Lee, EK(2017) PPtreeViz: An R Package for Visualizing Projection Pursuit Classification Trees, Journal of Statistical Software.
Examples
data(iris)
tree <- ODT(Species ~ ., data = iris)
#> Warning: You are creating a tree for classification
tree
#>
#> =============================================================
#> Oblique Classification Tree structure
#> =============================================================
#>
#> 1) root
#> node2)# proj1*X < 0.16 -> (leaf1 = setosa)
#> node3) proj1*X >= 0.16
#> node4)# proj2*X < 0.52 -> (leaf2 = versicolor)
#> node5)# proj2*X >= 0.52 -> (leaf3 = virginica)
print(tree, projection = TRUE, cutvalue = TRUE)
#>
#> =============================================================
#> Oblique Classification Tree structure
#> =============================================================
#>
#> 1) root
#> node2)# proj1*X < 0.16 -> (leaf1 = setosa)
#> node3) proj1*X >= 0.16
#> node4)# proj2*X < 0.52 -> (leaf2 = versicolor)
#> node5)# proj2*X >= 0.52 -> (leaf3 = virginica)
#>
#> Projection coefficient in each node
#> -------------------------------------------------------------
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> proj1 0.0000 -0.2722 0.0000 0.9622
#> proj2 -0.2008 0.0000 0.9796 0.0000
#>
#> Cutoff values of each node
#> -------------------------------------------------------------
#> CutValue1 CutValue2
#> 0.1638 0.5223