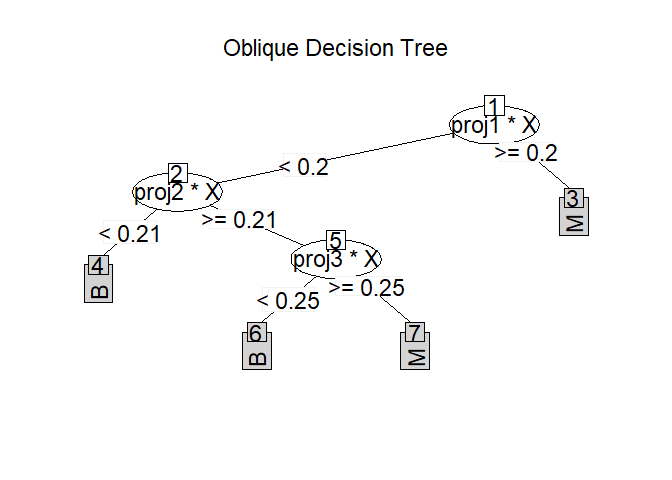
Prune ODRF from bottom to top with test data based on prediction error.
Usage
# S3 method for class 'ODRF'
prune(obj, X, y, MaxDepth = 1, useOOB = TRUE, ...)Arguments
- obj
An object of class
ODRF.- X
An n by d numeric matrix (preferable) or data frame is used to prune the object of class
ODRF.- y
A response vector of length n.
- MaxDepth
The maximum depth of the tree after pruning (Default 1).
- useOOB
Whether to use OOB for pruning (Default TRUE). Note that when
useOOB=TRUE,Xandymust be the training data inODRF.- ...
Optional parameters to be passed to the low level function.
Value
An object of class ODRF and prune.ODRF.
ppForestThe same result asODRF.pruneErrorError of test data or OOB after each pruning in each tree, misclassification rate (MR) for classification or mean square error (MSE) for regression.
Examples
# Classification with Oblique Decision Random Forest
data(seeds)
set.seed(221212)
train <- sample(1:209, 80)
train_data <- data.frame(seeds[train, ])
test_data <- data.frame(seeds[-train, ])
forest <- ODRF(varieties_of_wheat ~ ., train_data,
split = "entropy", parallel = FALSE, ntrees = 50
)
prune_forest <- prune(forest, train_data[, -8], train_data[, 8])
pred <- predict(prune_forest, test_data[, -8])
# classification error
(mean(pred != test_data[, 8]))
#> [1] 0.03875969
# \donttest{
# Regression with Oblique Decision Random Forest
data(body_fat)
set.seed(221212)
train <- sample(1:252, 80)
train_data <- data.frame(body_fat[train, ])
test_data <- data.frame(body_fat[-train, ])
index <- seq(floor(nrow(train_data) / 2))
forest <- ODRF(Density ~ ., train_data[index, ], split = "mse", parallel = FALSE, ntrees = 50)
prune_forest <- prune(forest, train_data[-index, -1], train_data[-index, 1], useOOB = FALSE)
pred <- predict(prune_forest, test_data[, -1])
# estimation error
mean((pred - test_data[, 1])^2)
#> [1] 5.565047e-05
# }